Modern Data Pipeline Implementation with PaasupDIP: Subway User Statistics Analysis Project #2
Introduction
In the previous blog post (Modern Data Pipeline with PaasUp DIP: Subway Passenger Analytics Project #1), we implemented a data pipeline by creating a Spark Session in a Jupyter Notebook environment.
In the second part of this series, we'll explore implementing and executing data pipelines in an actual production environment using Airflow, one of DIP's project service catalogs. We'll focus on workflow management and scheduling capabilities through Airflow DAGs.
Table of Contents
- 1. Project Overview
- 2. Airflow Catalog Deployment
- 3. Data Pipeline Modifications
- 4. Jupyter Notebook vs Airflow DAG: Spark Environment Comparison
- 5. Conclusion
1. Project Overview
Objectives
Moving beyond the development-stage Jupyter Notebook environment, we aim to implement the 'Subway Passenger Analytics Project' data pipeline in an Airflow production environment and build an automated workflow.
Technology Stack
- Data Processing: Apache Spark
- Data Storage: Delta Lake, PostgreSQL
- Visualization: Apache Superset
- Production Environment: Airflow
- Platform: DIP Portal
2. Airflow Catalog Deployment
-
Create Project Catalog
-
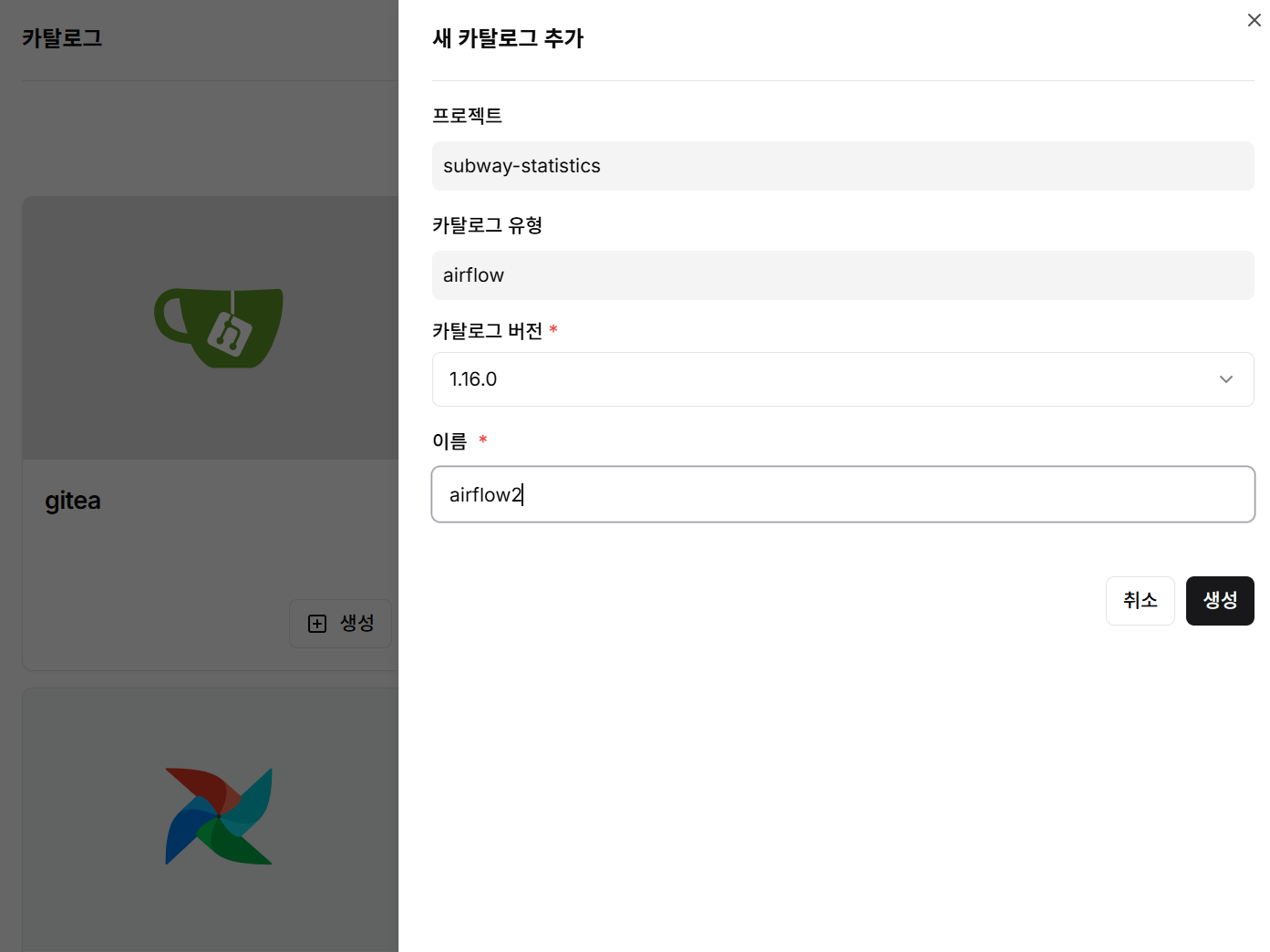
Select Airflow, define version and instance name, then deploy
-
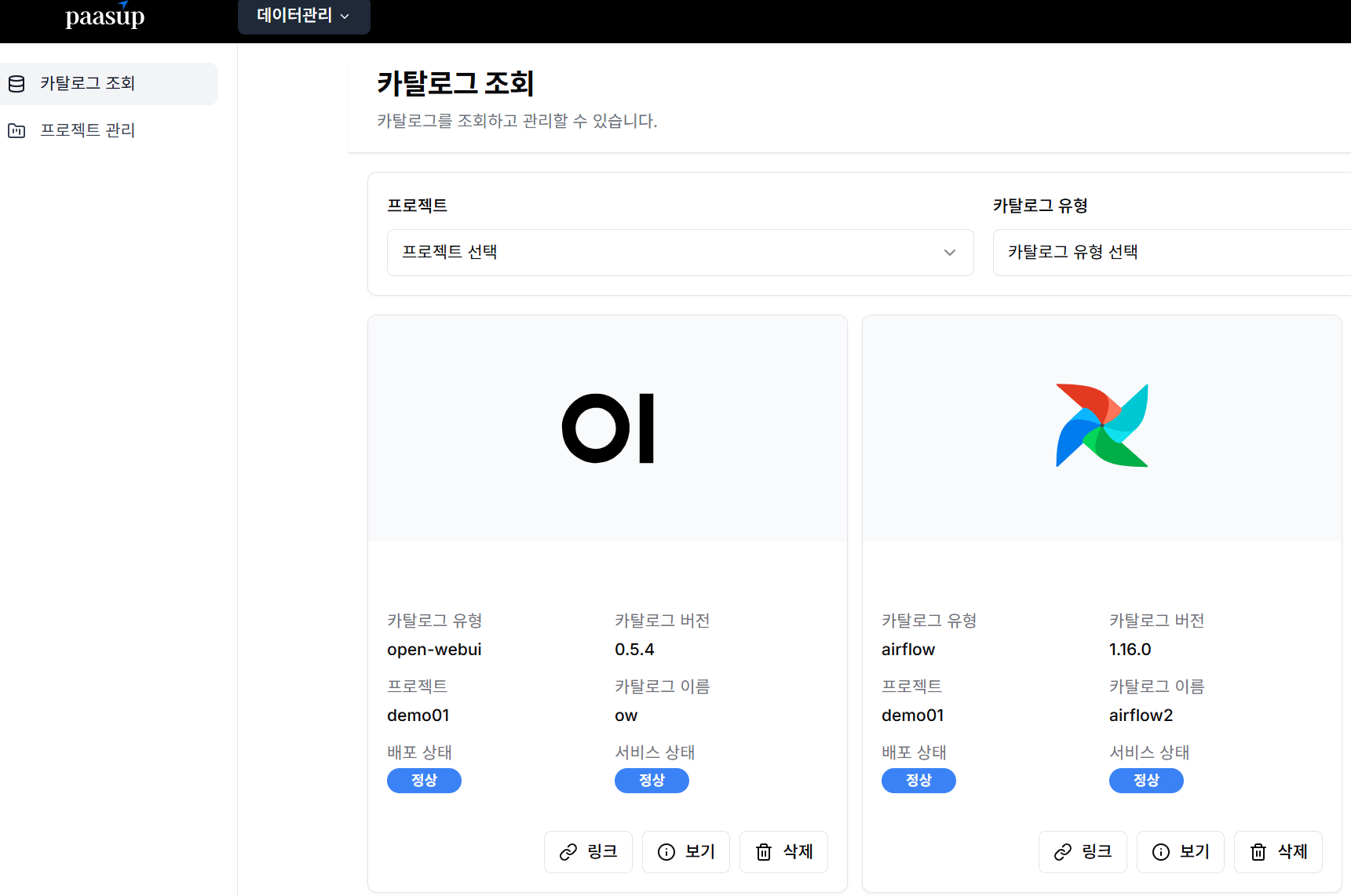
Query catalog to verify successful deployment
3. Data Pipeline Modifications
Environment Variable Configuration
In the Jupyter Notebook environment, we managed S3 and PostgreSQL connection information through .env files. However, in the Airflow production environment, we use Kubernetes Secrets for security and management efficiency. Sensitive connection information can be created as Secrets and safely referenced in spark-app.yaml.
$ export NAMESPACE="spark-job"
# Create S3 Secret
$ kubectl create -n $NAMESPACE secret generic s3-secret \
--from-literal=AWS_ACCESS_KEY_ID={your_s3_accesskey} \
--from-literal=AWS_SECRET_ACCESS_KEY={your_s3_secretKey} \
--from-literal=AWS_ENDPOINT_URL={your_s3_endpoint_url}
# Create PostgreSQL Secret
$ kubectl create -n $NAMESPACE secret generic pg-secret \
--from-literal=PG_HOST={your_pg_host} \
--from-literal=PG_PORT={your_pg_port} \
--from-literal=PG_DB={your_pg_dbname} \
--from-literal=PG_USER={your_pg_user} \
--from-literal=PG_PASSWORD={your_pg_password}
DAG File Creation
The Airflow DAG is a core component that defines the workflow structure and execution schedule. Here we configure a DAG for Spark jobs processing subway passenger data, designed to allow users to input the year-month data to be processed as parameters at runtime.
from datetime import datetime, timedelta
from airflow import DAG
from airflow.models.param import Param
from airflow.providers.cncf.kubernetes.operators.spark_kubernetes import SparkKubernetesOperator
from airflow.providers.cncf.kubernetes.sensors.spark_kubernetes import SparkKubernetesSensor
dag = DAG(
dag_id="load_subway_passengers",
default_args={'max_active_runs': 1},
description='Submit deltalake example as SparkApplication on Kubernetes',
schedule_interval=timedelta(days=1),
start_date=datetime(2025, 6, 1),
catchup=False,
# Define parameters at DAG level
params={
"WK_YM": Param(
default="202504",
type="string",
description="Work year-month (YYYYMM format)"
)
}
)
t1 = SparkKubernetesOperator(
task_id='load_subway_passengers',
namespace="demo01-spark-job",
application_file="./spark-app.yaml",
# Pass parameters through environment variables or template_fields
dag=dag
)
t1
Spark Application YAML Configuration
The SparkApplication YAML file is a resource definition for executing Spark jobs in a Kubernetes environment. This file includes Spark cluster configuration, resource allocation, environment variable settings, and configuration for dynamically fetching Python scripts from Git repositories.
apiVersion: "sparkoperator.k8s.io/v1beta2"
kind: SparkApplication
metadata:
name: load-subway-passengers-{{ params.WK_YM if params.WK_YM else data_interval_start.in_timezone("Asia/Seoul").strftime("%Y%m") }}
namespace: demo01-spark-job
spec:
type: Python
pythonVersion: "3"
mode: cluster
image: paasup/spark-offline:3.5.2-java17-python3.11-3
imagePullPolicy: IfNotPresent
mainApplicationFile: local:///git/dip-usecase/dags/load_subway_passengers/load_subway_passengers.py # Modify according to Git repository
sparkVersion: "3.5.2"
sparkConf:
spark.driver.extraJavaOptions: "-Divy.cache.dir=/tmp -Divy.home=/tmp -Dcom.amazonaws.sdk.disableCertChecking=true -Duser.timezone=Asia/Seoul"
spark.executor.extraJavaOptions: "-Dcom.amazonaws.sdk.disableCertChecking=true -Duser.timezone=Asia/Seoul"
spark.hadoop.fs.s3a.endpoint: "https://172.16.50.29:9000"
spark.hadoop.fs.s3a.impl: "org.apache.hadoop.fs.s3a.S3AFileSystem"
spark.hadoop.fs.s3a.path.style.access: "True"
spark.hadoop.fs.s3a.connection.ssl.enabled: "True"
spark.hadoop.fs.s3a.aws.credentials.provider: "EnvironmentVariableCredentialsProvider"
spark.sql.extensions: "io.delta.sql.DeltaSparkSessionExtension"
spark.sql.catalog.spark_catalog: "org.apache.spark.sql.delta.catalog.DeltaCatalog"
spark.kubernetes.driverEnv.WK_YM: "{{ params.WK_YM if params.WK_YM else data_interval_start.in_timezone("Asia/Seoul").strftime("%Y%m") }}"
# No need for below configuration as jar files are included in Docker image
# Even if jar files exist in the image, adding below configuration attempts download, so exclude when files are in the image
spark.jars.packages: "org.apache.hadoop:hadoop-common:3.3.4,org.apache.hadoop:hadoop-aws:3.3.4,com.amazonaws:aws-java-sdk-bundle:1.12.262,io.delta:delta-spark_2.12:3.3.1,org.postgresql:postgresql:42.7.2"
timeToLiveSeconds: 120
restartPolicy:
type: Never
volumes:
- name: git-volume
emptyDir: {}
driver:
cores: 1
coreLimit: "1200m"
memory: "2048m"
labels:
version: 3.5.2
serviceAccount: spark
volumeMounts:
- mountPath: /git
name: git-volume
# initContainer to fetch Python scripts for Spark execution from airflow-dags repository
initContainers:
- name: "init-clone-repo"
image: bitnami/git:2.40.1-debian-11-r2
imagePullPolicy: IfNotPresent
command:
- /bin/bash
args:
- -ec
- git clone https://github.com/paasup/dip-usecase.git --branch main /git/dip-usecase # Configure according to Git repository
volumeMounts:
- mountPath: /git
name: git-volume
#envFrom:
#- secretRef:
#name: git-secret # Needs to be created. Exclude for public repositories
envFrom:
- secretRef:
name: s3-secret # Create S3 or MinIO connection information to use
- secretRef:
name: pg-secret # Create PostgreSQL connection information to use
executor:
instances: 1
cores: 1
memory: "2g"
labels:
version: 3.5.2
Python Script Modifications
The data pipeline code written in the existing Jupyter Notebook needs to be modified to be executable in the Airflow environment. The main changes include execution environment detection, improved parameter handling, and file format conversion.
1. Execution Environment Detection Function Implementation
To use the same code in both development and production environments, we implement a function that detects whether the current execution environment is Jupyter Notebook. This allows applying different parameter processing logic based on the environment.
def is_running_in_jupyter():
try:
# Check if running in IPython environment
shell = get_ipython().__class__.__name__
if shell == 'ZMQInteractiveShell':
return True # Jupyter Notebook or QtConsole
elif shell == 'TerminalInteractiveShell':
return False # IPython running in terminal
else:
return False # Other environments
except NameError:
return False # Not IPython (regular Python interpreter)
2. Improved Parameter Processing Logic
Modify the main execution block to handle parameters differently based on the execution environment. In Jupyter environment, use hardcoded values for testing, and in Airflow environment, dynamically receive parameters from environment variables.
# --- Main execution block ---
if __name__ == "__main__":
if is_running_in_jupyter():
WK_YM = '202504'
else:
# input parameters by airflow
WK_YM = os.environ['WK_YM']
print("------------------------------")
print("WK_YM : " + WK_YM)
print("-------------------------------")
3. Notebook File Format Conversion
Convert Jupyter Notebook files (.ipynb) to regular Python script files (.py) for direct execution in Airflow and Spark. During this process, the notebook's cell structure is removed, leaving only pure Python code.
Airflow Job Registration and Execution
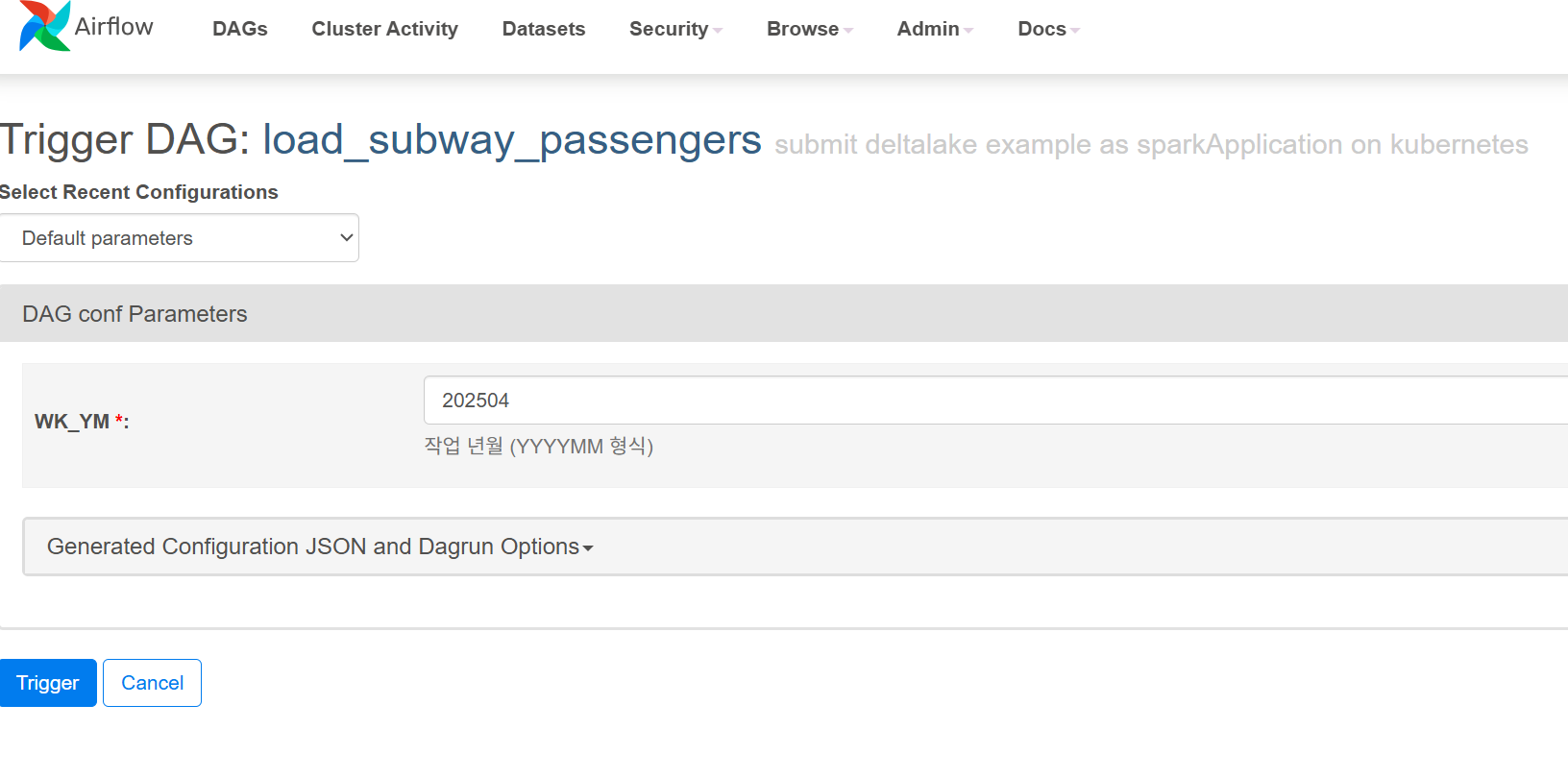
Airflow jobs are automatically registered through the Git repository information defined in the initContainers of spark-app.yaml. When you click the execution button for the corresponding job in the Airflow web portal, a screen appears where you can input parameters to pass to the Spark Application. After entering the required parameters, you can execute the job by clicking the Trigger button.
The job execution process is as follows:
- DAG files are automatically detected from the Git repository and registered in Airflow
- Select job and click execution button in web UI
- Set required values in parameter input screen (e.g., year-month data to process)
- Execute job and monitor through Trigger button
4. Jupyter Notebook vs Airflow DAG: Spark Environment Comparison
Using Spark in Jupyter notebook environment versus Airflow DAG environment has significant differences across multiple aspects.
Execution Environment Differences
Jupyter Notebook Environment
- Interactive environment with cell-by-cell code execution
- Developers directly write and test code in real-time through browser
- Immediate result verification and easy debugging
- Spark Context remains alive while session is maintained
Airflow DAG Environment
- Batch processing environment with scheduled job execution
- Code is packaged as Python scripts and executed on worker nodes
- Executed as part of automated workflows
- New Spark session is created and terminated for each task
Key Characteristics Comparison
| Characteristic | Jupyter Notebook | Airflow DAG |
|---|---|---|
| Resource Management | • Manual resource management by developers • Possible resource occupation due to long-running sessions • Can monopolize cluster resources |
• Automatic resource release after task completion • Dynamic resource allocation possible • Efficient resource sharing among multiple DAGs |
| Error Handling & Retry | • Manual error handling • Developer manually re-executes on failure • Detailed information available for debugging |
• Automatic retry mechanism • Notifications and logging on failure • Dependency management and backpressure handling |
| Scheduling & Dependencies | • Manual execution • Sequential execution by default • No dependency management |
• Cron-based scheduling • Complex dependency graph management • Parallel execution possible |
| Monitoring & Logging | • Result confirmation through cell output • Limited logging • Real-time monitoring difficult |
• Centralized logging • Monitoring through web UI • Job history and metrics tracking |
| Security & Access Control | • Personal development environment • Possibility of hardcoded credentials in notebooks • Risk of security information exposure during version control |
• Safe credential management through Connections and Variables • RBAC-based access control • Security model suitable for production environments |
5. Conclusion
Jupyter is suitable for development and prototyping, while Airflow is suitable for operating automated data pipelines in production environments.
The typical development pattern is to validate logic with Jupyter during the development phase, then convert it to Airflow DAGs for production operation.
